Protein Structure Analysis
Structural and dynamic analysis of proteins is an emerging field of biological mass spectrometry and complements information which can be obtained by NMR, cryo-EM and other technologies.
With the development of Biological MS instruments, software and methods it is now possible to obtain detailed information about disordered protein regions, folding, protein-protein interactions, and protein small molecule interactions.
We apply cross-linking MS (XL-MS) and hydrogen deuterium MS (HDX) to gain insights in structures and dynamics of proteins and protein complexes.
Support within protein structure analysis at BioMS include:
High quality data from our HDX-MS platform comprised of a LEAP-PAL3 autosampler custom built for HDX sample preparation, and an Ultimate 3000 micro-LC system coupled to an Orbitrap MS. The automated sample handling is essential to provide the, reproducibility, and the measurement precision needed for analysis of sample series.
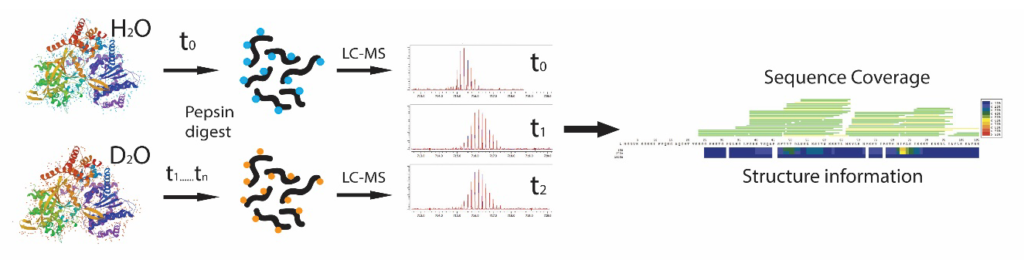
A typical HDX-MS experiment entails H/D exchange by incubating the sample in buffer with deuterated water during defined time-points, proteolytic digestion at low pH, LC-MS separation of peptides, followed by data analysis.
Cross linking (XL-MS) experiments with a variety of cross linkers including MS cleavable reagents are offered. In-solution or in-gel digested cross linked samples are analyzed with Fusion MS instrument. Enrichment of cross-linked peptides is performed with size exclusion chromatography for complex samples.
Contact

