Chemical Proteomics
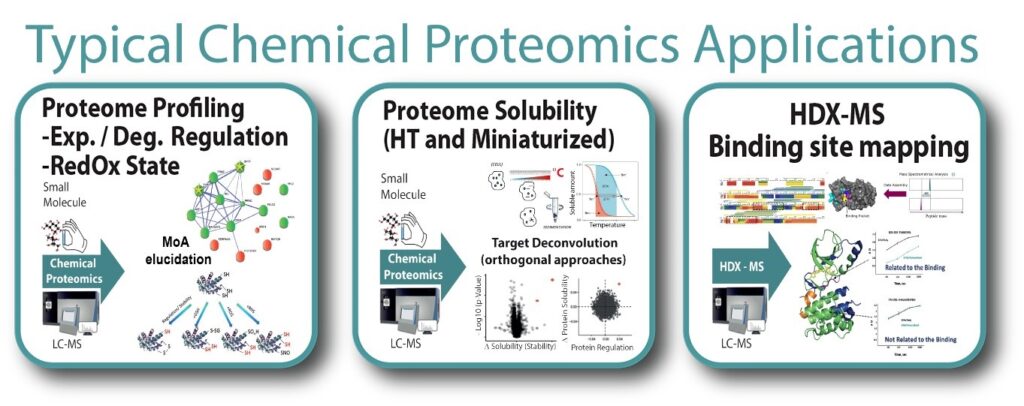
Chemical proteomics includes a powerful set of complementary, mass spectrometry(MS)-based approaches for identifying the key proteins relevant for producing the phenotypic effects of a treatment with a molecule of a potential new drug, including discovery of protein direct and indirect targets as well as mechanism of action in a proteome-wide manner.
The paradigm in chemical proteomics using modified drugs as baits for activity- and affinity-based approaches using chemically engineered ligand probes, has recently shifted to the use of proteome-wide approaches with no ligand modification, for the uncertainty in the functional properties of the engineered bait compared to the original ligand and for the limited the analysis specificity after the use of the bait. However, as complementary tool to proteome-wide chemical proteomics provided at our node, quantitative proteomics after affinity-based approaches is also provided.
Our Chemical Proteomics node early adopted several state of-of-the-art chemical proteomics methods for target deconvolution, such as Thermal Proteome Profiling (TPP), and successfully developed and optimized a new method for target discovery and mechanism of action elucidation named PISA (Proteome Integral Solubility Alteration assay) that overcomes potential pitfalls of previous methods providing altogether much higher levels (>15 folds compared to TPP) of throughput, sustainability and statistical reliability, by making use of several biological replicates multiplexed in one experiment, together with the potential use of different ligands, e.g. active and inactive drugs / compounds of interest.
Such approaches wide-open the accessibility of several projects to chemical proteomics and allow integration with other methods to reach multidimension proteome profiling analysis, with maintaining high sustainability and final proteome analysis depth. Moreover, PISA dramatically reduces sample requirement and miniaturization down to primary cultures, organoids and iPSCs systems is today feasible in chemical proteomics.
The use of orthogonal approaches dramatically increases confidence in common hits defining key proteins in the mechanism of action, and for this reason quantitative proteomics on the global proteome amount regulation after treatment is also utilized orthogonally for evaluating the specific regulation of certain proteins due to a certain treatment, as in the methods FITExP and ProTargetMiner (the last for anticancer molecules).
Another dimension of chemical proteomics under development and currently being optimized is RedOX proteomics, to identify oxidative changes on protein cysteines at the peptide level. This dimension is also thought as integration of the above.
Moreover, the binding site of a chemical compound to a particular protein can be probed by deuterium exchange experiments using mass spectrometry.
Support within chemical proteomics at BioMS include:
- High throughput, thermally challenged proteome solubility profiling for deconvolution of targets, off-targets and elucidation of mechanism of action (PISA, TPP, integrated PISA with orthogonal approaches)
- Identification of specific protein regulation of targets using multiple molecular proteome signatures (FITExP, ProTargetMiner for anticancer molecules)
- RedOx Proteomics
- PTMs analysis related to the mechanism of action investigated
- Protein ID and relative quantification after affinity-based approaches
- Identification of enzyme substrates using PISA
- Interaction interface mapping of protein targets of potentially therapeutic small molecules and antibodies (epitope mapping) using Hydrogen / Deuterium exchange mass spectrometry (HDX-MS)
Contact
Massimiliano Gaetani

